Слайд 2
Genome, the entire genetic complement of an organism
Genomics,
research that addresses all or a substantial portion of
an organism’s genome
Includes physical mapping & sequencing of all or a large part of a genome or chromosome
Слайд 3
Why Study Genomes of Different Organisms?
To understand the
genetics behind diseases (Homo sapien
& Canis familiarus)
To learn more about human pathogens & how to prevent or treat their infections (Clostridium tetani, Bacillus anthacis, & Haemophilus influenzae)
Understand & improve the genetics of commercial organisms (Lactococcus lactis, Oryza sativa, Bos taurus, & Gallus gallus)
To discover the workings of unusual or odd organisms (Bdellovibrio bacteriovorus & Deinococcus radiodurans)
To understand phyolegeny
Слайд 4
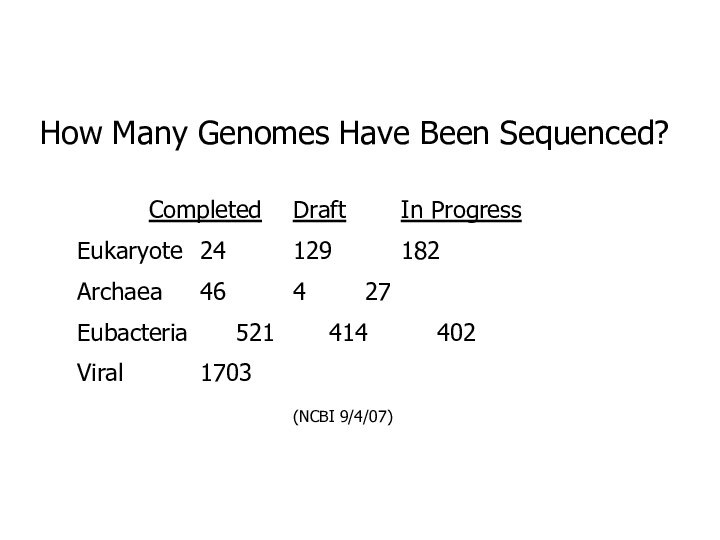
How Many Genomes Have Been Sequenced?
Completed Draft In Progress
Eukaryote 24 129 182
Archaea
46 4 27
Eubacteria 521 414 402
Viral 1703
(NCBI 9/4/07)
Слайд 5
How Do We Measure a Genome?
1 base=1
nucleotide=1basepair (bp)
1000bases=1kilobase (Kb)
1000kb=1megabase (Mb)
1000mb=1gigabase (Gb)
Слайд 6
Genome Sizes (haploid)
Organism Genome in Mb
E. coli 4.64
Yeast 12
Nematode 97
Fruit Fly 170
Pufferfish 345
Human 3200
Lungfish 129000
Слайд 7
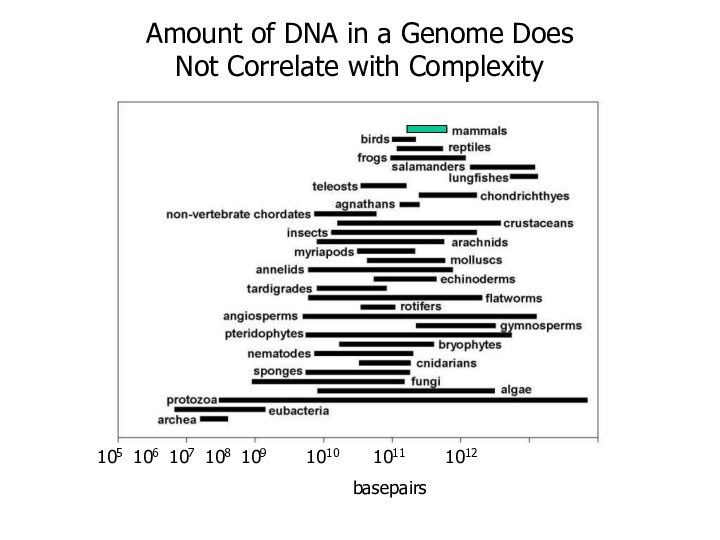
105 106 107 108 109 1010 1011
1012
basepairs
Amount of DNA in a Genome
Does Not Correlate with Complexity
Слайд 8
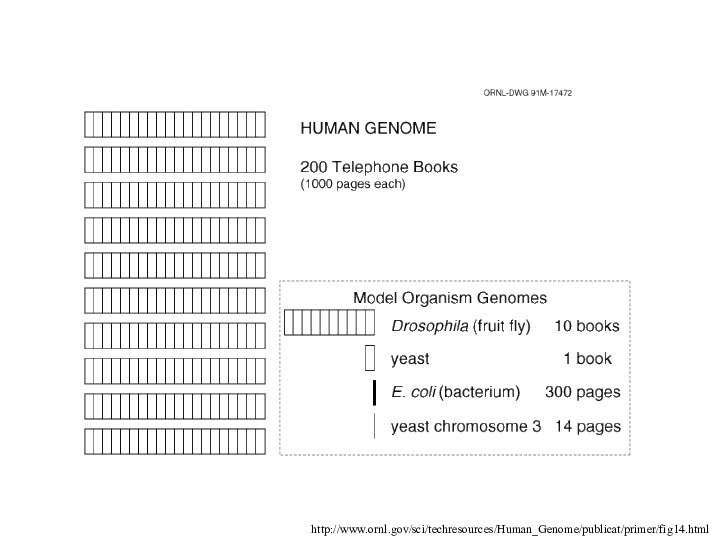
http://www.ornl.gov/sci/techresources/Human_Genome/publicat/primer/fig14.html
Слайд 9
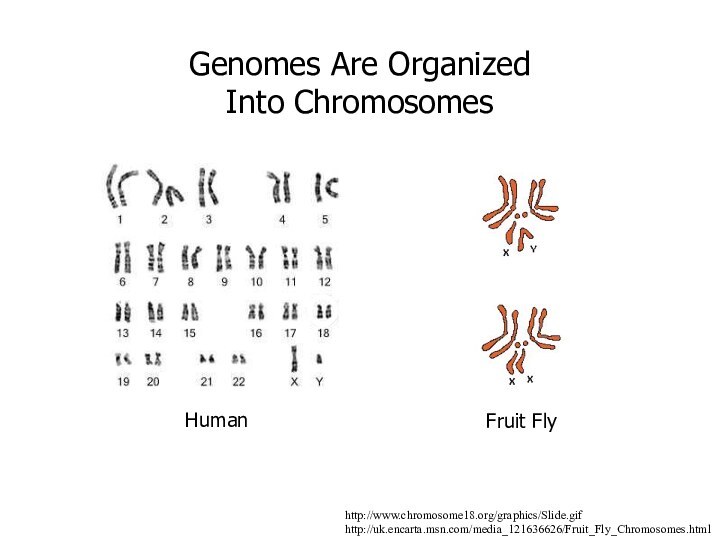
http://www.chromosome18.org/graphics/Slide.gif
http://uk.encarta.msn.com/media_121636626/Fruit_Fly_Chromosomes.html
Genomes Are Organized Into Chromosomes
Human
Fruit Fly
Слайд 10
Chromosome Number Is Species Specific
Diploid Number 2n
Human 46
Mouse 40
Fruit Fly 8
Dog 78
Arabidopsis 10
Corn 20
Yeast 32
Crayfish 200
Слайд 11
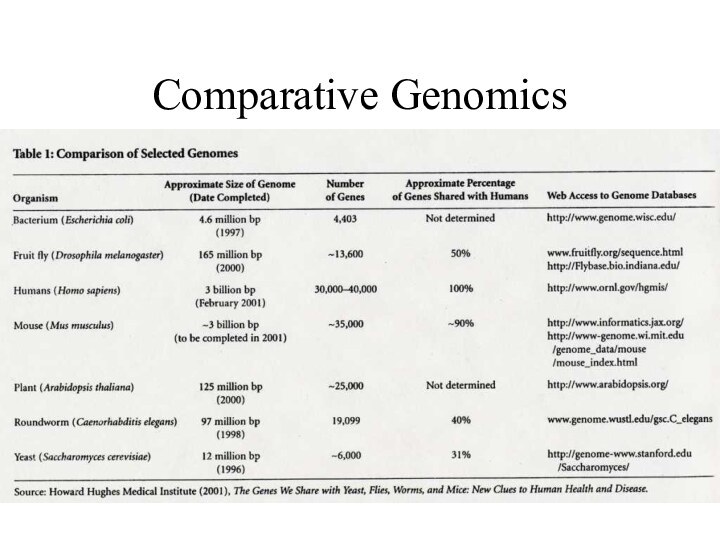
How many genes do we have?
Original estimate was
between 50 000 to 100 000 genes
We now think
human have ~ 25 000 genes
How does this compare to other organisms?
Mice have ~30 000 genes
Pufferfish have ~35 000 gene
The nematode (C. elegans), has ~19 000
Yeast (S. cerevisiae) there are ~6000 genes
The microbe responsible for tuberculosis has ~4000
Слайд 12
http://www.mun.ca/biology/desmid/brian/BIOL2250/Week_Two/genespac.jpg
Gene Spacing in Various Species
Слайд 13
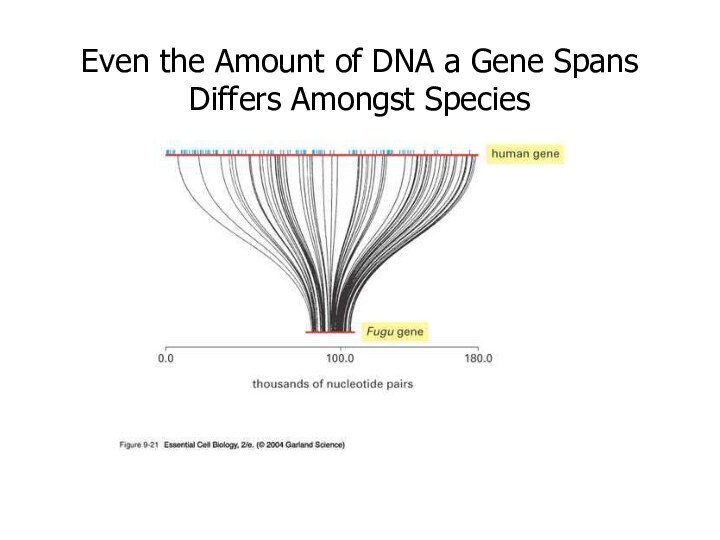
Even the Amount of DNA a Gene Spans
Differs Amongst Species
Слайд 15
Yeast
70 human genes are known to repair
mutations in yeast
Nearly all we know about cell cycle
and cancer comes from studies of yeast
Advantages:
fewer genes (6000)
few introns
31% of yeast genes give same products as human homologues
Слайд 16
Drosophila
nearly all we know of how mutations
affect gene function come from Drosophila studies
We share 50%
of their genes
61% of genes mutated in 289 human diseases are found in fruit flies
68% of genes associated with cancers are found in fruit flies
Knockout mutants
Homeobox genes
Слайд 17
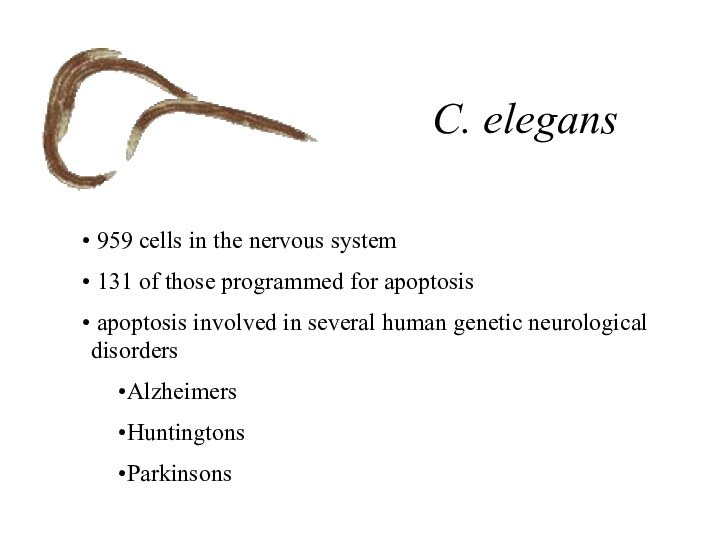
C. elegans
959 cells in the nervous system
131 of those programmed for apoptosis
apoptosis involved in
several human genetic neurological disorders
Alzheimers
Huntingtons
Parkinsons
Слайд 18
Mouse
known as “mini” humans
Very similar physiological systems
Share
90% of their genes
Слайд 19
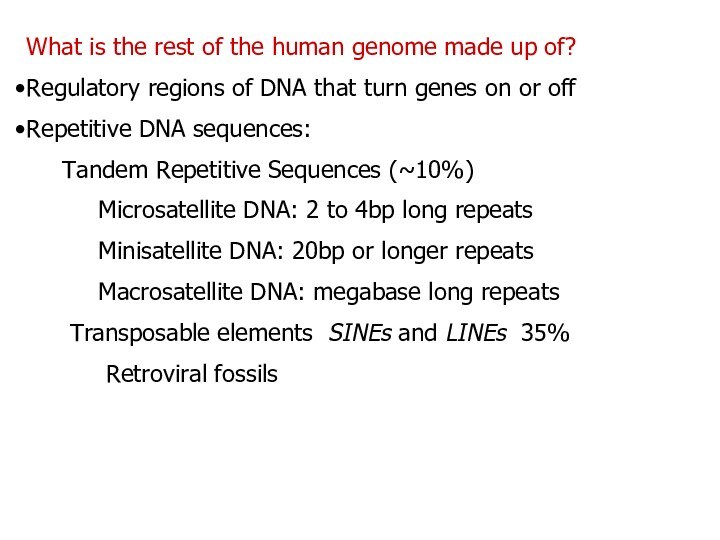
What is the rest of the human genome
made up of?
Regulatory regions of DNA that turn genes
on or off
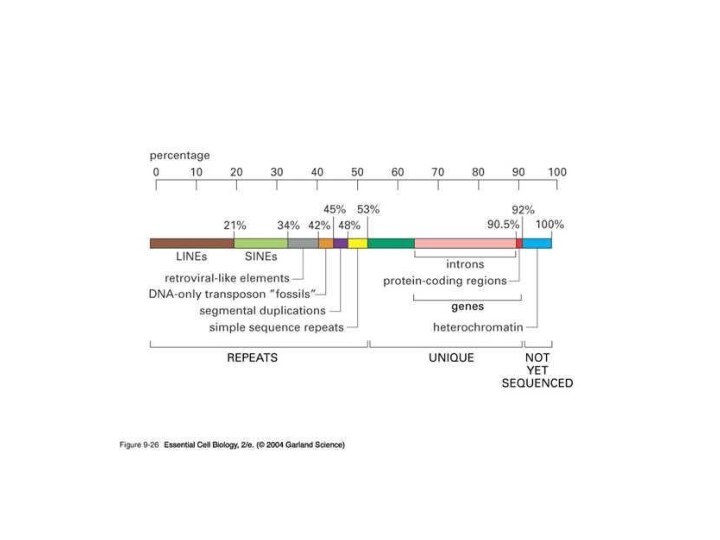
Repetitive DNA sequences:
Tandem Repetitive Sequences (~10%)
Microsatellite DNA: 2 to 4bp long repeats
Minisatellite DNA: 20bp or longer repeats
Macrosatellite DNA: megabase long repeats
Transposable elements SINEs and LINEs 35%
Retroviral fossils
Слайд 22
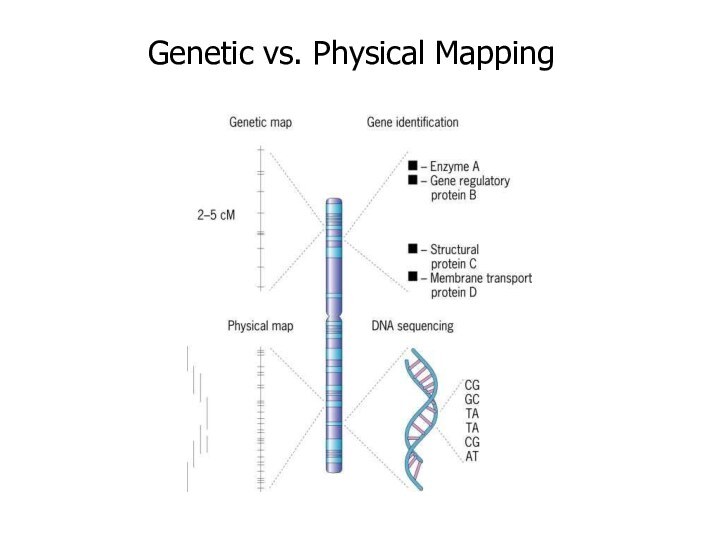
Genetic mapping based on genetic techniques, maps show
the positions of diseases or traits based on recombination
frequencies
Genetic techniques include cross-breeding experiments or, the examination of family histories (pedigrees)
Physical mapping uses molecular biology techniques to examine DNA molecules directly to construct maps showing the positions of sequence features, including genes
Physical techniques include DNA restriction enzyme analysis & fluorescent tagging of chromosomal regions
Слайд 23
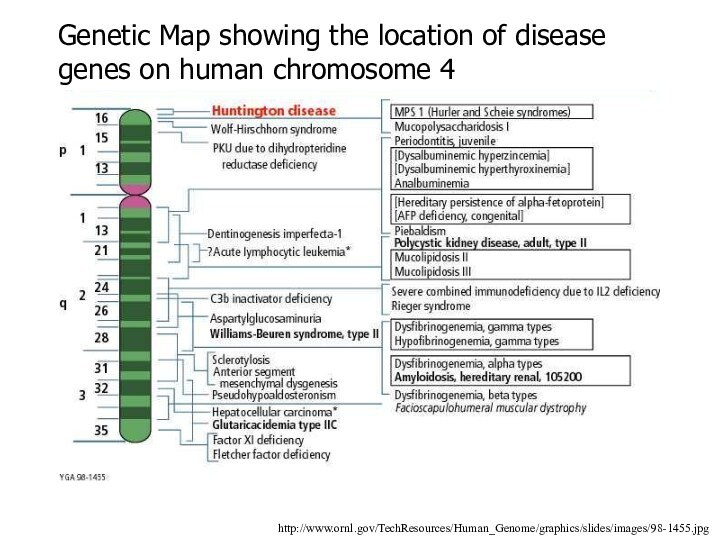
http://www.ornl.gov/TechResources/Human_Genome/graphics/slides/images/98-1455.jpg
Genetic Map showing the location of disease genes
on human chromosome 4
Слайд 24
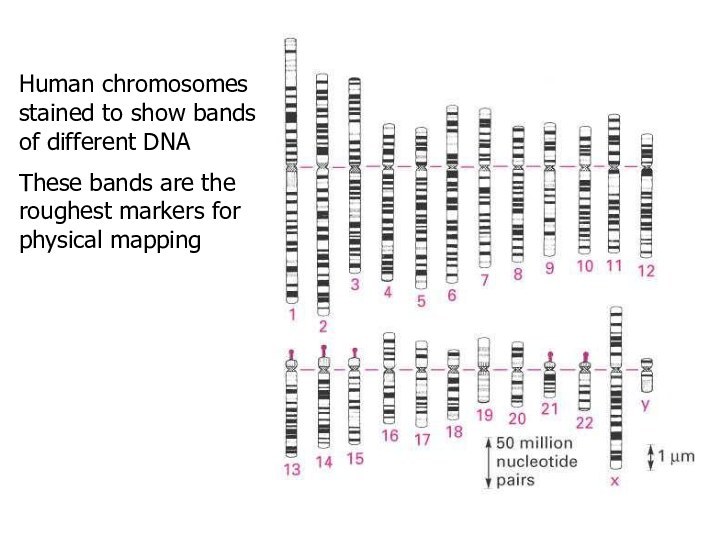
Human chromosomes stained to show bands of different
DNA
These bands are the roughest markers for physical mapping
Слайд 25
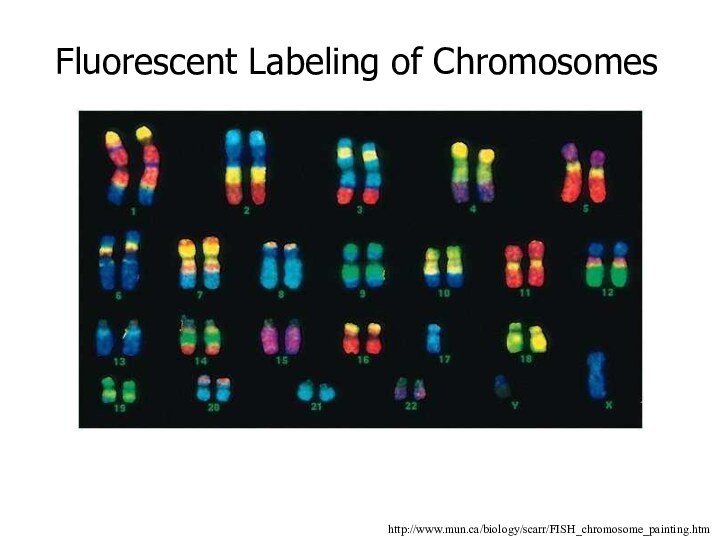
Fluorescent Labeling of Chromosomes
http://www.mun.ca/biology/scarr/FISH_chromosome_painting.htm
Слайд 26
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=hmg.figgrp.1556
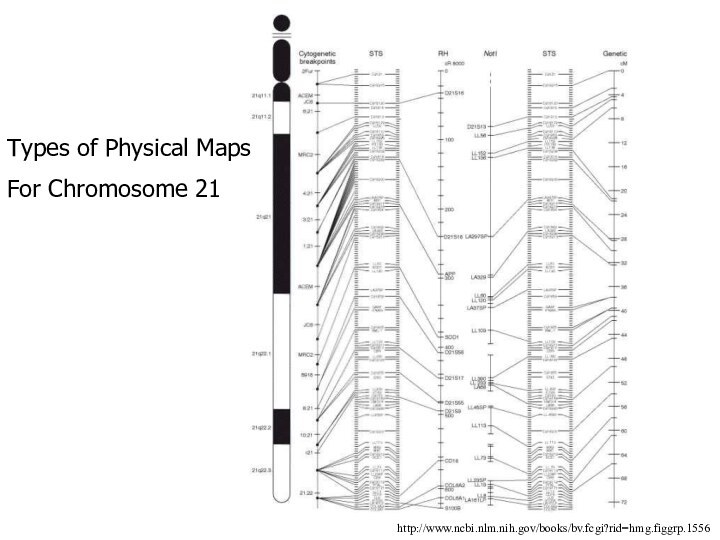
Types of Physical Maps
For Chromosome 21
Слайд 27
http://freepages.genealogy.rootsweb.com/~patafordgenealogy/images/usmaphyperlinks.jpg
http://socialstudies.ccswebacademy.net/CivicsEconomicsJenkins/images/Map_Outline_US_Outline.jpg
The more markers better the resolution, the more
useful the map
Слайд 29
Polyacrylamide gel electrophoresis can resolve ssDNA molecules that
differ in length by just one nucleotide
A banding pattern
is produced after separation of ssDNA molecules by denaturing polyacrylamide gel electrophoresis
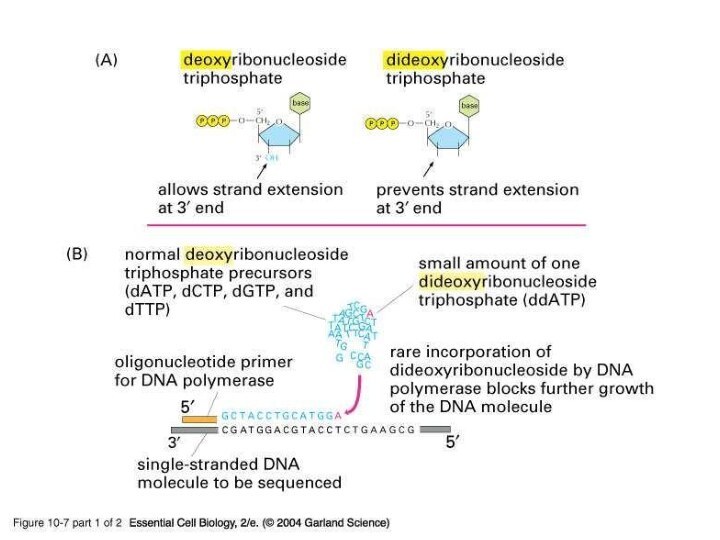
Слайд 31
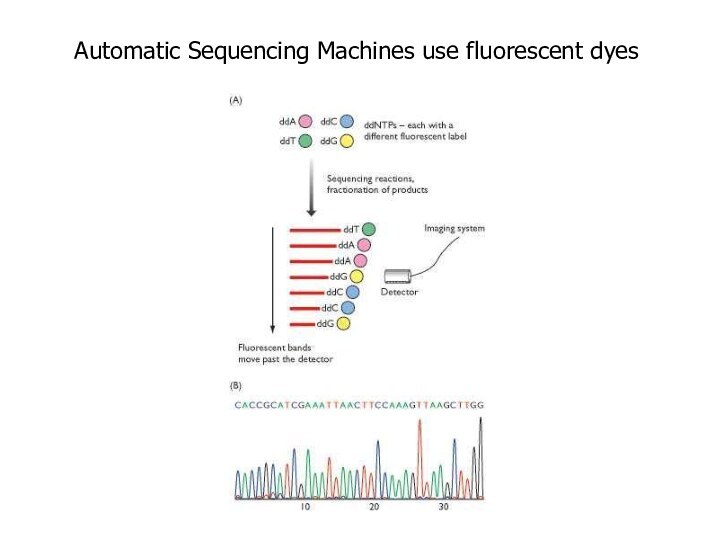
Automatic Sequencing Machines use fluorescent dyes
Слайд 32
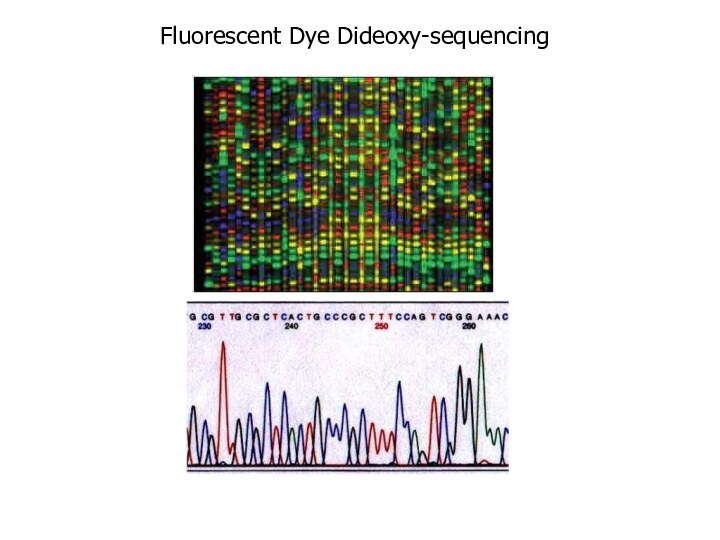
Fluorescent Dye Dideoxy-sequencing
Слайд 33
http://www.ornl.gov/TechResources/Human_Genome/graphics/slides/ttseqfacility.html
DNA Sequencers in Action
Слайд 34
First Complete Sequence of a Free-Living Organism
Слайд 35
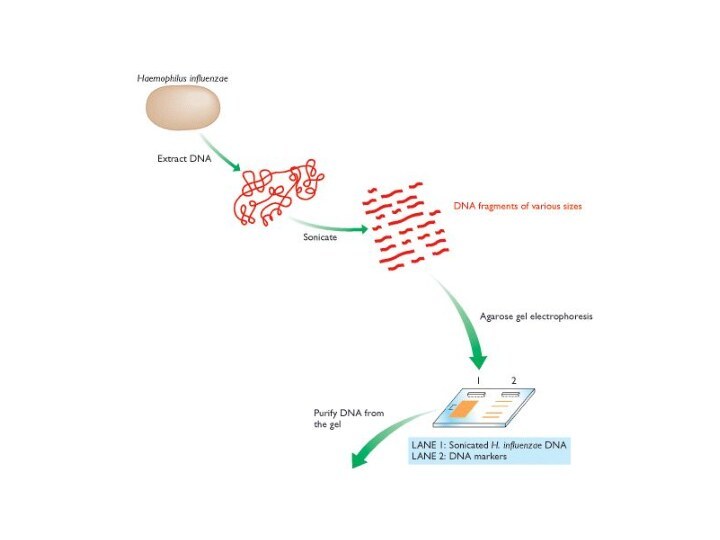
1995, the Haemophilus influenzae genome sequenced
Genome size=1830 kb
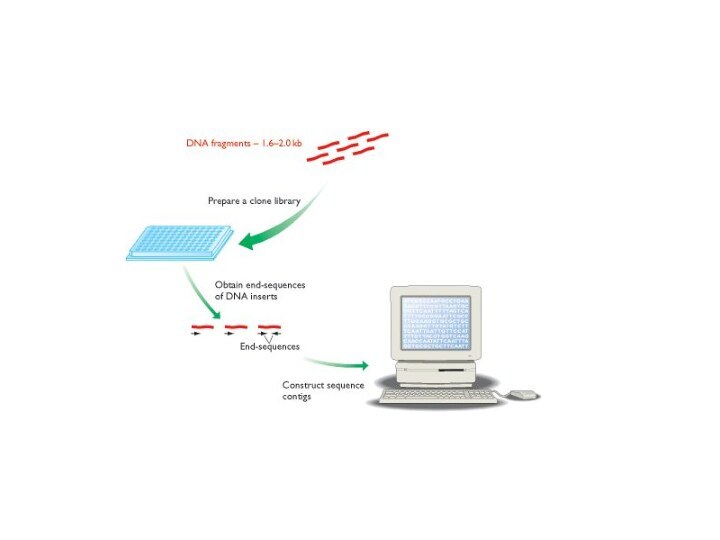
1st genome sequenced using the shotgun method
28,643 sequencing experiments
totaling 11,631,485 bp
This equaled 6x the length of the H. influenzae genome
Sequence assembly 30 hrs on a computer with 512 Mb of RAM
Resulted in 140 lengthy contiguous sequences
Each sequence contig represented, non-overlapping portion of the genome
Слайд 39
1st proposed by the DoE 1984
By 1990,
the Human Genome Project was launched
The Human Genome
Organization (HUGO) was founded to provide a forum for international coordination of genomic research
The program was proposed to include:
The creation of genetic & physical maps to be used in the generation of a complete genome sequence
Human Genome Project
Слайд 40
First Steps of the Human Genome Project
1) Construct
genetic & physical maps of the haploid human &
mouse genomes
These would provide key tools for identification of disease genes and anchoring points for genomic sequence
2) Sequence the yeast and worm genomes, as well as targeted regions of mammalian genomes
Слайд 41
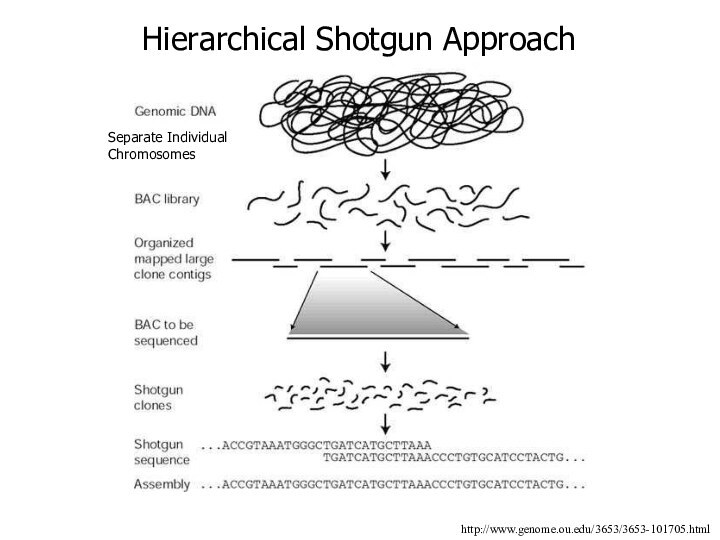
Sequencing Plan of HUGO
1) Isolate each human chromosome
2)
Physical mapping of each chromosome
The banding pattern of visible
through staining
Location of known genes already mapped
Location of restriction enzyme sites
Chromosome fragmented into large pieces of DNA and inserted into BAC or YAC libraries
Fragments overlap such that they can be ordered into a rough assembly of the chromosome
DNA from 5 humans
2 males, 3 females
2 caucasians, one each of asian, african, hispanic
Слайд 42
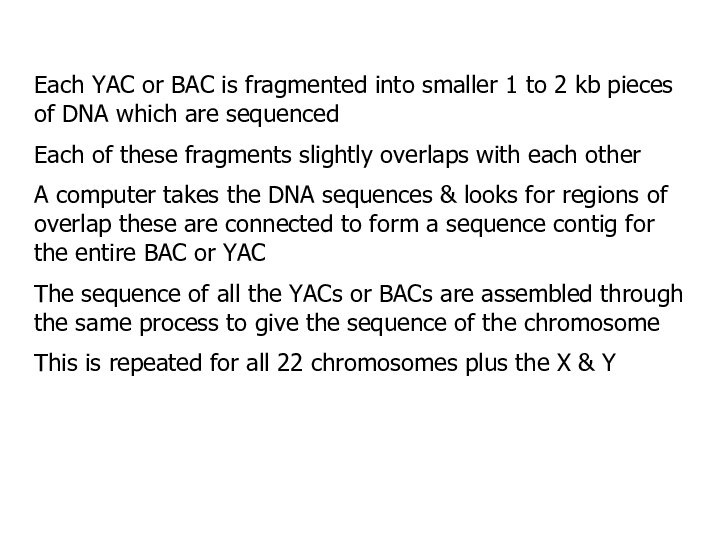
Each YAC or BAC is fragmented into smaller
1 to 2 kb pieces of DNA which are
sequenced
Each of these fragments slightly overlaps with each other
A computer takes the DNA sequences & looks for regions of overlap these are connected to form a sequence contig for the entire BAC or YAC
The sequence of all the YACs or BACs are assembled through the same process to give the sequence of the chromosome
This is repeated for all 22 chromosomes plus the X & Y
Слайд 43
Hierarchical Shotgun Approach
http://www.genome.ou.edu/3653/3653-101705.html
Separate Individual Chromosomes
Слайд 45
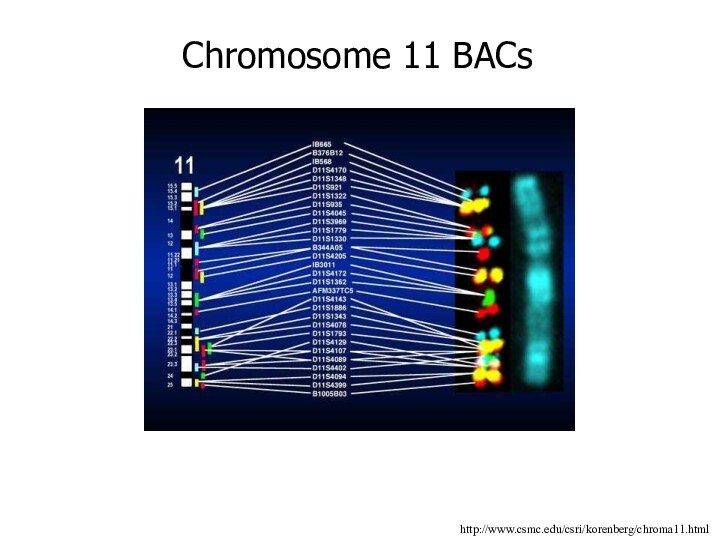
http://www.csmc.edu/csri/korenberg/chroma11.html
Chromosome 11 BACs
Слайд 46
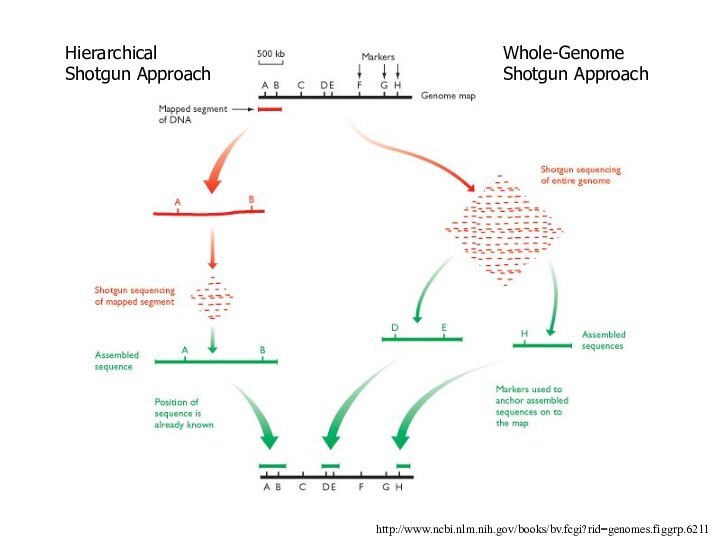
1999, Celera Genomics, set out to sequence the
human genome using a whole-genome shotgun method - more
riskier - goal to patent some seq.
There would be no isolation of individual chromosomes & no subcloning into BACs or YACs
They skipped straight to the 1 to 2 kb fragments
The $300 million Celera effort was intended to proceed at a faster pace and at a fraction of the cost of the roughly $3 billion HUGO project.
Dr. Craig Venter (founder) Celera Genomics
Human Genome Whole-Genome Shotgun Method
Слайд 47
14.8-billion bp of DNA sequence was generated over
9 months
This equaled 5x the human genome
Resulting sequence contigs
spanned >99% of the genome
In March 2000, President Clinton announced that the genome sequences could not be patented, and should be made freely available to all researchers. The statement sent Celera's stock plummeting.
The competition proved to be very good for the project, spurring the public groups to modify their strategy in order to accelerate progress.
In February 2001 Celera Genomics published their draft of the human genome in the journal Science
The same month HUGO published its draft of the human genome in the journal Nature
The rivals initially agreed to pool their data, but the agreement fell apart when Celera refused to deposit its data in the unrestricted public database GeneBank.
Слайд 48
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=genomes.figgrp.6211
Hierarchical Shotgun Approach
Whole-Genome Shotgun Approach
Слайд 49
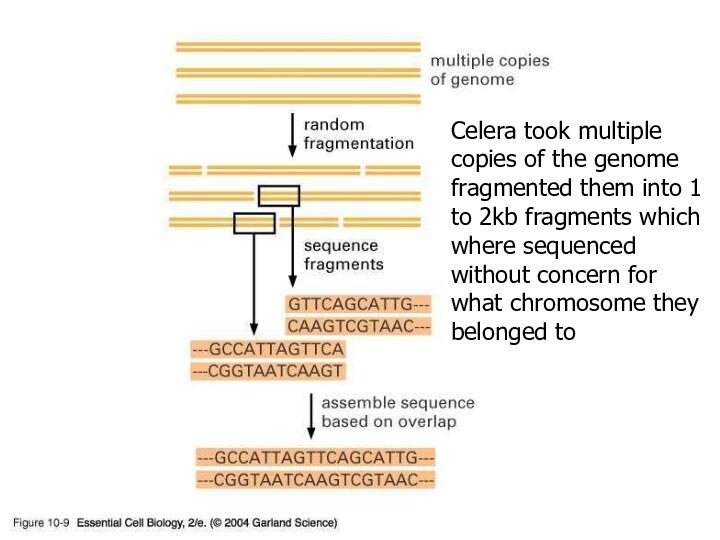
Celera took multiple copies of the genome fragmented
them into 1 to 2kb fragments which where sequenced
without concern for what chromosome they belonged to
Слайд 50
What did they learn?
1.1% of the genome is
spanned by exons
24% is in introns
75% of the genome
is intergenic DNA
A random pair of human haploid genomes differs on average at a rate of 1 bp per 1250 bp
Слайд 51
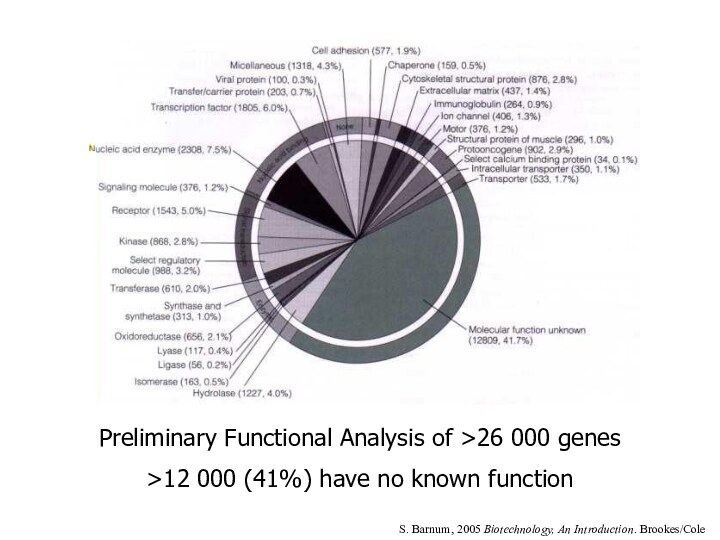
Preliminary Functional Analysis of >26 000 genes
>12 000
(41%) have no known function
S. Barnum, 2005 Biotechnology, An
Introduction. Brookes/Cole
Слайд 52
Diploid Genome Sequence of an Individual Human
On September
4th, 2007, a team led by Craig Venter, published
his (ovn) complete DNA sequence, unveiling the six-billion-letter genome of a single individual for the first time.
44% of known genes had one or more alterations
>0.5% variation between two haploid genomes
Слайд 53
How Do We Differ?
Total of 4.1 million DNA
variations
3.2 million single nucleotide changes
53,800 block substitutions (2 to
206bp)
292,000 heterozygous insertion/deletions (1 to 571bp)
559,000 homozygous insertion/deletions (1 to 82,711bp)
90 inversions
Numerous duplications & copy number variations
Слайд 55
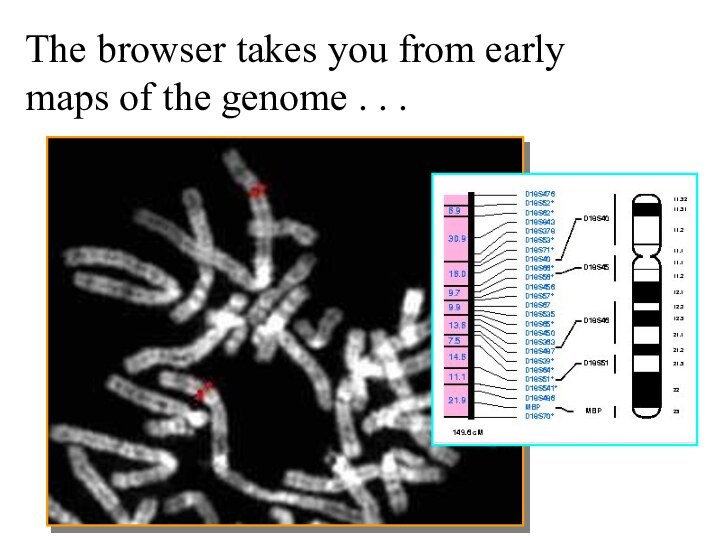
The browser takes you from early maps of
the genome . . .
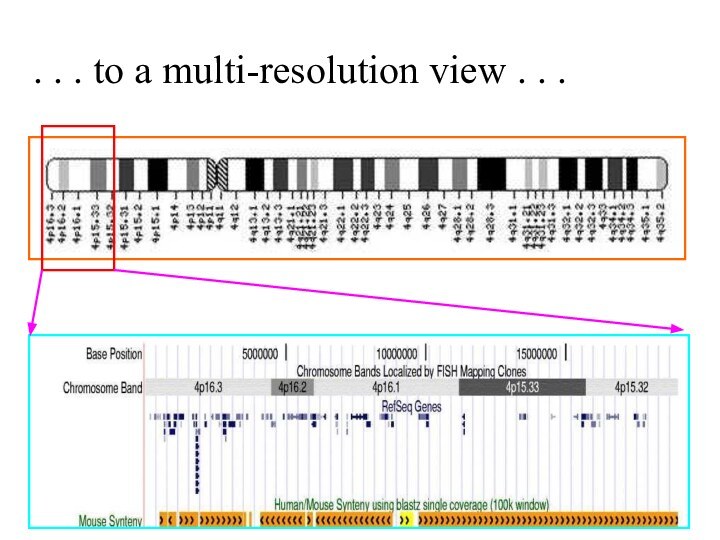
Слайд 56
. . . to a multi-resolution view .
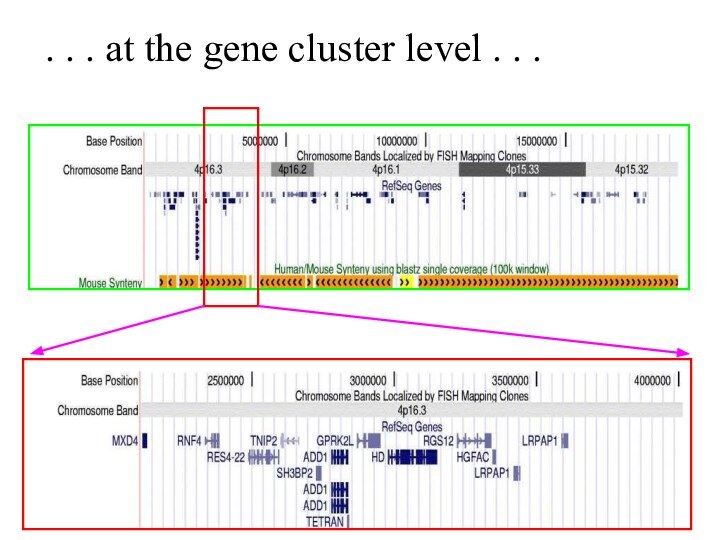
Слайд 57
. . . at the gene cluster level
. . .
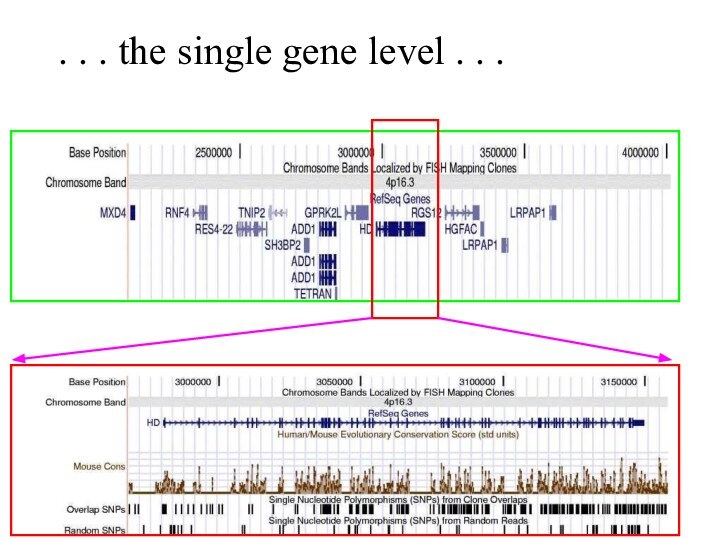
Слайд 58
. . . the single gene level .
. .
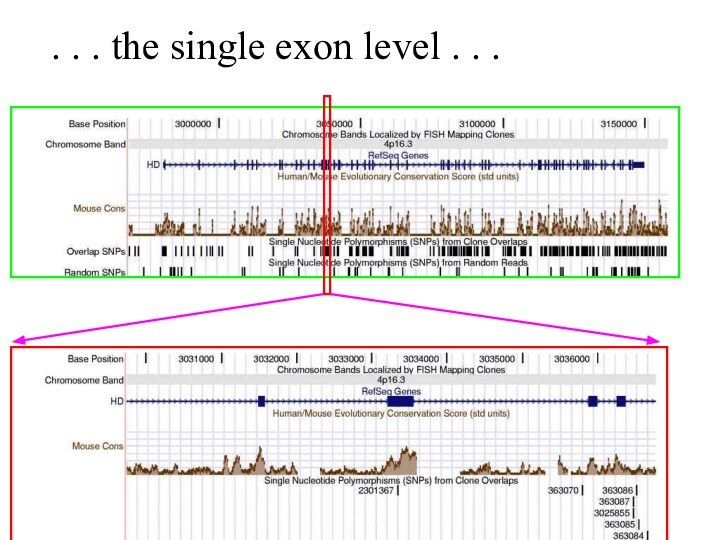
Слайд 60
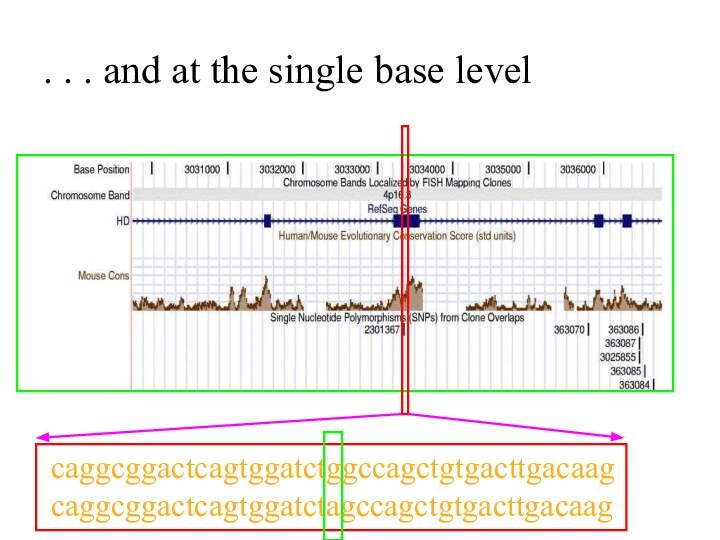
. . . and at the single base
level
caggcggactcagtggatctggccagctgtgacttgacaag
caggcggactcagtggatctagccagctgtgacttgacaag